DeepEvidence
Deep Knowledge Graph Research Agent
DeepEvidence is a hierarchical multi-agent system for comprehensive biomedical literature research and evidence synthesis. It leverages deep knowledge graph exploration to systematically gather, analyze, and synthesize evidence from multiple biomedical knowledge bases.
Multi-Agent Architecture
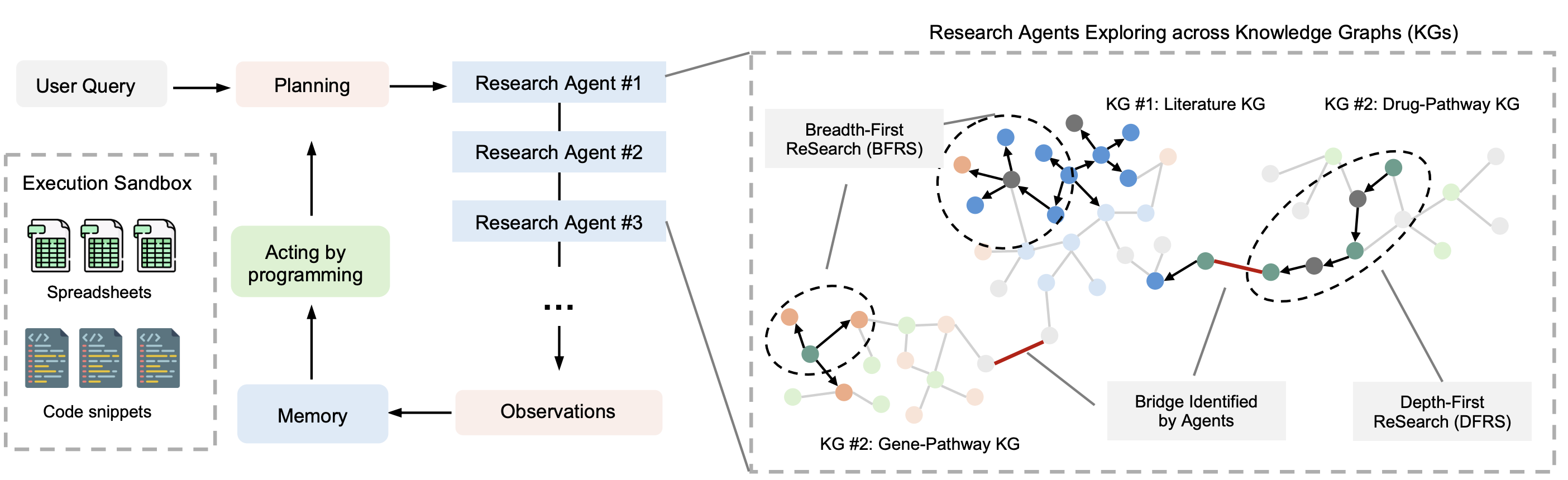
Orchestrator Agent
Coordinates research strategy, decides which knowledge bases to explore, and synthesizes findings.
BFRS Agent
Breadth-First ReSearch (BFRS) of knowledge graphs to discover related concepts and broad connections.
DFRS Agent
Depth-First ReSearch (DFRS) of specific knowledge paths to extract detailed information.
Unified Knowledge Graph APIs
DeepEvidence integrates 15+ biomedical APIs across multiple domains for comprehensive research.
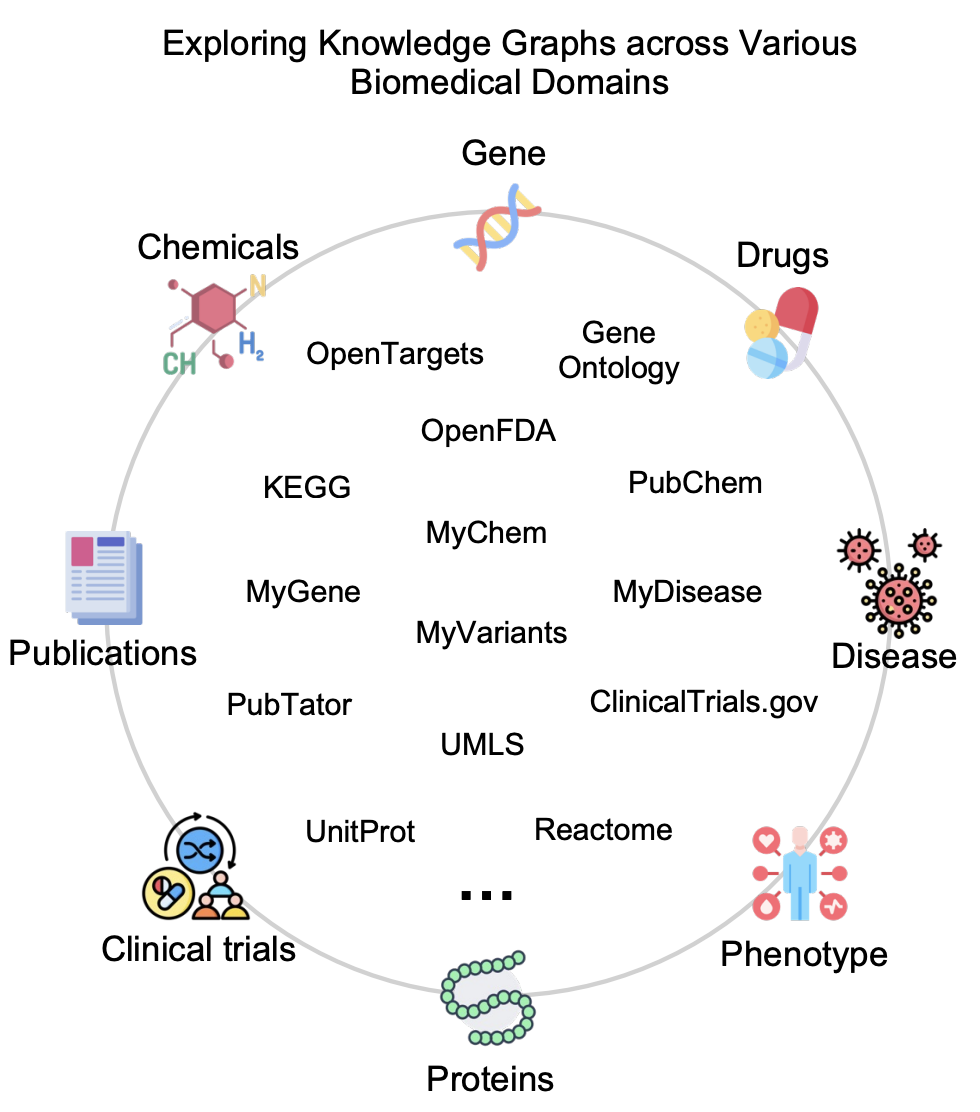
Literature & Publications
Genes & Proteins
Drugs & Chemicals
Diseases & Phenotypes
Pathways & Reactions
Clinical Data
Evidence Graph
DeepEvidence builds a persistent knowledge graph during research that captures entities (papers, genes, diseases, drugs) and their relationships.
- Accumulates knowledge across search rounds
- Enables retrieval of previously discovered information
- Supports iterative refinement of research questions
- Exports to interactive HTML/PDF visualizations
# Export evidence graph
results.export_evidence_graph_html(
"evidence_graph.html"
)
# Access discovered entities
entities = results.evidence_graph_data[
'entities'
]
relations = results.evidence_graph_data[
'relations'
]Evidence Graph Exploration
Interactive visualization showing how DeepEvidence iteratively builds a knowledge graph through systematic exploration. Use the controls to step through the 6 phases of target identification.
Benchmark Results
DeepEvidence significantly outperforms existing methods across biomedical research benchmarks.
HLE-Medicine
Hard medicine questionsLabBench-LitQA2
Literature QASuperGPQA-Hard
Expert medicine questionsTrialPanorama
Evidence synthesisDeepEvidence Benchmark
7 knowledge-graph-driven deep research tasks spanning the biomedical discovery pipeline.
Target Identification
25 tasksIdentify therapeutic targets for diseases by integrating gene-disease associations and pathway evidence.
MoA Pathway Reasoning
25 tasksMulti-hop mechanistic reasoning to explain molecular perturbation propagation through pathways.
Metabolic Flux Response
25 tasksPredict metabolic flux suppression in pre-clinical models based on pathway dependencies.
Drug Regimen Design
25 tasksDesign drug dosing regimens considering pharmacological and clinical factors.
Surrogate Endpoint
14 tasksIdentify plausible surrogate endpoints that reflect downstream clinical outcomes.
Sample Size Estimation
25 tasksEstimate clinical trial sample sizes under design assumptions and outcome constraints.
Evidence Gap Discovery
20 tasksIdentify missing, weak, or conflicting evidence across biomedical knowledge sources.
Example Discovery Tasks
DeepEvidence tackles complex biomedical research questions requiring multi-hop reasoning across knowledge bases.
Quick Start
from biodsa.agents import DeepEvidenceAgent
# Initialize the agent
agent = DeepEvidenceAgent(
model_name="gpt-5",
api_type="azure",
api_key=os.environ.get("AZURE_OPENAI_API_KEY"),
endpoint=os.environ.get("AZURE_OPENAI_ENDPOINT"),
model_kwargs={
"max_completion_tokens": 5000,
"reasoning_effort": "minimal",
},
subagent_action_rounds_budget=5, # action rounds for sub research agents
main_search_rounds_budget=2, # search rounds for main orchestrator
main_action_rounds_budget=15, # action rounds for main orchestrator
light_mode=False, # use full memory graph
llm_timeout=120,
)
# Run the agent
execution_results = agent.go(
"Summarizing the cutting-edge immunotherapy drugs in late clinical "
"trial phase or have been approved for NSCLC?",
knowledge_bases=[
"pubmed_papers", "clinical_trials", "drug", "disease"
]
)Citation
@article{wang2025deepevidence,
title = {DeepEvidence: Empowering Biomedical Discovery with Deep Knowledge Graph Research},
author = {Wang, Zifeng and Chen, Zheng and Yang, Ziwei and Wang, Xuan and Jin, Qiao and Peng, Yifan and Lu, Zhiyong and Sun, Jimeng},
journal = {arXiv preprint},
year = {2025}
}